Running a PyTorch machine learning model on an ESP32
I wanted to figure out how to run a PyTorch machine learning model on an ESP32. I use MicroPython pretty often so would be even better if I could do it natively on MicroPython.
I didn't really care what the model did so I chose handwriting detection as an arbitrary test case. There is a PCB with a bunch of pads that I use as touch sensors and then use my finger to draw alphabets on them and try and get the model to guess which letter I drew.
Couple things I find cool about this:
Model was trained on PyTorch on my laptop and inference performed on an ESP32
The way this is implemented it would be quite easy to completely change the model architecture and reconfigure it to do other things like audio recognition or object detection
The ML related custom code-base is extremely small (~120 lines of MicroPython code and ~235 lines of C optimizations)
Good enough for real-time inference (needs 31ms for inference)
Has 95% validation set accuracy (as a reference point - a simpler technique like measuring distance from the mean of each category got only 31% accuracy)
Hardware
ESP32 connected to a display
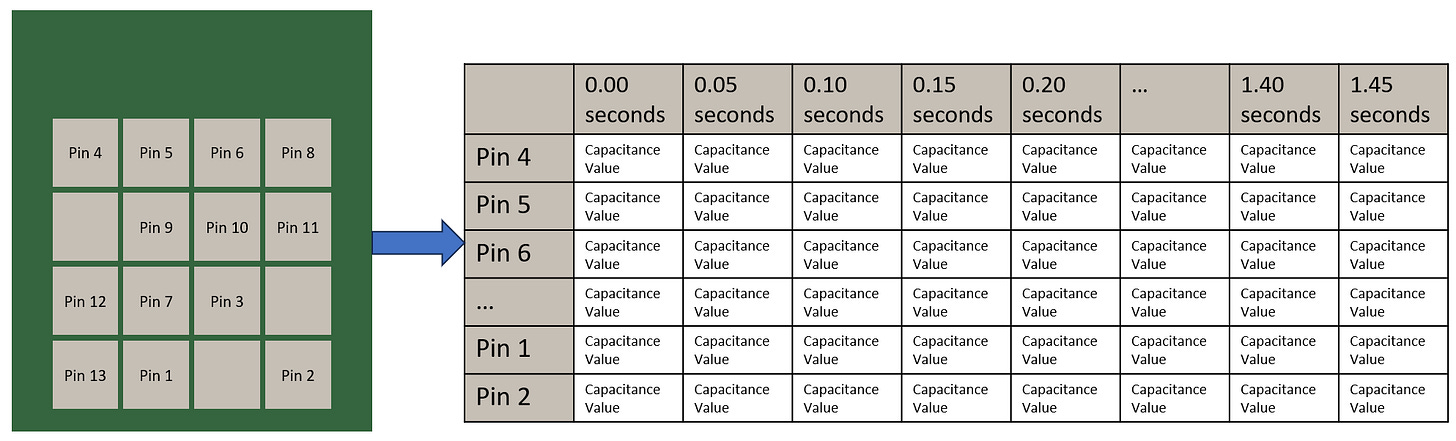
Pads on a PCB that act as touch sensors
Only 13 of the 16 pads are connected due to MCU peripheral limitations - the MCU only has 13 capacitive touch input input pins
USB for power
Data
To collect training data I used my finger to draw out specific shapes for each letter and stored that data onto the ESP
Each "gesture" has touch sensing data from each of the pads collected at 50ms intervals for 1.5s
The data array is 13 sensors x 30 datapoints, each datapoint being 50ms apart.
Since this is a 2D array of floats, the techniques that apply to image classification models will also apply here.
To see why, let's visualize the data. Below is what one sample of each 1.5s "recording" of the input data looks like when visualized for each letter. The more green something is, the higher the capacitance i.e. the stronger the touch detection signal (larger contact area will lead to larger signal amplitude)
Data is stored onto the ESP32 as a CSV file then transferred onto desktop for training
I collected 120 samples of each letter which takes about 3 minutes of writing the same letter onto the pad each time (Note: Since I had to manually record the data I have only implemented detection for the alphabets A, B, C, X, Y, and Z. Doing all 26 alphabets would be around 1.5 hours of just writing alphabets onto the touchpad - not necessary for a proof of concept on a for-fun blog post)
Training - PyTorch / Laptop
Here is the model architecture I landed on after trying a couple.
| model = nn.Sequential( | |
| ### 13 x 30 input | |
| ### 2 stride, 3x3 kernel, 3 output channel convolution layer | |
| ### = 3 channels x 6 rows x 14 columns output | |
| nn.Conv2d(1, 3, (3,3), 2), | |
| ### Activation layer | |
| nn.ReLU(), | |
| ### Dropout to help generalize | |
| nn.Dropout(0.1), | |
| ### 2 stride, 3x3 kernel, 12 output channel convolution layer | |
| ### = 12 channels x 2 rows x 6 columns output | |
| nn.Conv2d(3, 12, (3,3), 2), | |
| ### Activation layer | |
| nn.ReLU(), | |
| ### Dropout to help generalize | |
| nn.Dropout(0.1), | |
| ### 1 stride, 2x3 kernel, 12 output channel convolution layer | |
| ### = 12 channels x 1 row x 4 columns output | |
| nn.Conv2d(12, 12, (2,3), 1), | |
| ### Activation layer | |
| nn.ReLU(), | |
| ### Dropout to help generalize | |
| nn.Dropout(0.1), | |
| ### Reshape data from 12 x 1 x 4 tensor -> 48 long vector | |
| nn.Flatten(), | |
| ### Linear layer with 48 inputs and 7 outputs | |
| ### One output each for: | |
| ### no gesture detected / A / B / C / X / Y / Z) | |
| nn.Linear(48,7) | |
| ) |
This model has 1585 parameters in total and requires 11,287 floating point operations.
I wouldn't be surprised if this architecture can be optimized further to have fewer parameters (reduce storage requirements) and reduce the number of floating point operations (run faster by reducing the amount of computation required)
It got 95% validation set accuracy but in real life testing there's still plenty of room for improvement because it occasionally confuses one letter for another or is not confident enough about the letter it predicts
I should collect more training data with more varied ways of writing the letters and that will probably improve the model. eg. B needs to be written a specific way because that's how I ended up writing it while I was recording training data.
This model took 13 minutes to train on my laptop with an RTX4060 to get to 95% accuracy (I could get to around 90% accuracy by training for 2.5 minutes)
I won't get too into the details of how the model was trained - that's not really the focus of this article. The key thing to note is there is a model that takes 13 x 30 matrix of touch sensor data as input and can predict which of the 6 alphabets it is with 95% accuracy (in theory - in practice it is worse).
The default way to store the weights for a PyTorch models is a .pt file but since we want to write our own weights loader in micropython it will be a lot easier to save the weights as a JSON file. Luckily this is pretty easy and can be done with:
with open(f'{fname}.json', 'w') as json_file:
json.dump(model.state_dict(), json_file,cls=EncodeTensor)We will look at how to load these into our MicroPython model later but first - we need to actually make the MicroPython model.
Inference - MicroPython / ESP32
To recreate the model for inference on the ESP32 here's what we need to do:
Recreate the different model layers (
nn.Conv2d,nn.Linear,nn.ReLU,nn.Flatten)Write a weights loader that can read the JSON weights file and load the weights into the model layers from step 1
Performance optimizations to speedup inference (optional, of course)
Validate results against running the model with same data on desktop to ensure same output
Creating different blocks
I really just needed 5 types of layers -
nn.Conv2d,nn.Linear,nn.ReLU,nn.Flatten, and a dummy layer that does nothing to simulate thenn.Dropoutlayers that don't do anything during inference and only make a difference during training.
I first wrote all the code in pure MicroPython so that I could validate my logic and find out which bits of code were slow and needed to be sped up using C.
I'm going to show how the most complicated layer (the convolution layer) works below since I want to show that even that isn't really a lot of code and is actually quite easy to implement. At the end of the article I'll include all the layers as an appendix so anyone that wants to can have a look.
For those who don't care to read the code (I get it!) here's a quick walkthrough showing you what's being done. Note that I'm going to assume you already know what a convolution is - if not, check this out (link). Alternatively for the purposes of this article you can think of it like this: a convolution takes in a 2D array as an input and applies a function to it and gives back another 2D array as an output. The function can be something like edge detection and the output can be a 2D array where all edges are white and any pixel that isn't an edge is black.
Back to the layer implementation - an actual convolution layer can have multiple input channels but I've shown a simplified case with one input channel and multiple output channels below to keep things easy to understand. With one input channel what a convolution layer does is apply n_out_channel number of kernels (functions) to the input "image". As an example - if I had a convolution layer that had 3 output channels and the channels detected left edges, top edges, and outlines then this is what that would look like.

The first layer of our model also has one input and 3 outputs but the kernels seem more complicated than just edge detection. Here is what the first convolution layer of the model I trained actually does. I could take a guess as to what it might be doing but honestly I'm not confident so let's leave that out for now. There are techniques to figure what the different layers are doing but that's probably a whole blog post in and of itself.
In code, the nn.Conv2d equivalent is:
| class convLayer(Module): | |
| def __init__(self, in_channels, out_channels, kernel_size: tuple, stride): | |
| self.kernels = [[None for _ in range(in_channels)] for _2 | |
| in range(out_channels)] | |
| self.bias = [None for _ in range(out_channels)] | |
| self.stride = stride | |
| self.kernel_size = kernel_size | |
| def __call__(self, images): | |
| conv_func = convolution2D | |
| output_data = [] | |
| for out_channel in range(len(self.kernels)): | |
| channel_output = [] | |
| for in_channel, image in enumerate(images): | |
| channel_output.append(conv_func(image, self.kernels[out_channel][in_channel],self.stride)) | |
| ### Matrix addition operation | |
| ### sum up all the different outputs for each channel | |
| summed_channel_output = sumMatrices(channel_output) | |
| ### Add the bias for each channel to each element | |
| ### in the output matrix for that channel | |
| biased_channel_output = addBias(summed_channel_output, self.bias[out_channel]) | |
| output_data.append(biased_channel_output) | |
| return output_data | |
| def convolution2D(image, kernel, stride): | |
| ### Convolve single 2D matrix with kernel using stride | |
| ### returns a single 2D matrix | |
| ### Get dimensions of the input matrix (image) and the kernel | |
| image_height, image_width = int(len(image)), int(len(image[0])) | |
| kernel_height, kernel_width = int(len(kernel)), int(len(kernel[0])) | |
| ### Calculate dimensions of the output image | |
| output_height = (image_height - kernel_height) // stride + 1 | |
| output_width = (image_width - kernel_width) // stride + 1 | |
| ### Initialize output image with zeros | |
| output = [[0]*output_width for _ in range(output_height)] | |
| ### Iterate over the image | |
| for i in range(0, image_height, stride): | |
| ### optimization | |
| output_y = i//stride | |
| for j in range(0, image_width, stride): | |
| ### optimization | |
| output_x = j//stride | |
| ### Check if the kernel can fit in the image | |
| if i + kernel_height <= image_height and j + kernel_width <= image_width: | |
| ### Apply the kernel to the image | |
| for m in range(kernel_height): | |
| i_plus_m = i+m | |
| for n in range(kernel_width): | |
| output[output_y][output_x] += image[i_plus_m][j+n] * kernel[m][n] | |
| return output |
Similarly I've implemented the other layers from my PyTorch model and you can have a look at the code in the appendix. To put all of them together I have a Model class. Let's look at how it works first then we can look at how it's implemented.
This is how you'd use the Model class.
| layers_pred = [convLayer(1, 3, (3,3), 2), | |
| ### 3 x 6 x 14 | |
| ReLU(), | |
| dummyLayer(), | |
| convLayer(3, 12, (3,3), 2), | |
| ### 3 x 2 x 6 | |
| ReLU(), | |
| dummyLayer(), | |
| convLayer(12, 12, (2,3), 1), | |
| ### 12 x 1 x 4 | |
| ReLU(), | |
| dummyLayer(), | |
| Flatten(), | |
| linearLayer(48,7)] | |
| model_name = "model_weights.json" | |
| pred_model = Model(layers_pred, 35887.27,9239.95).load_weights(model_name) # The two numbers are the mean and standard deviation of the data and they are used to normalize data before feeding it into the model |
And this is how it's implemented - pretty simple.
| class Model: | |
| def __init__(self, layers, inp_norm_mean=0, inp_norm_sd=1): | |
| self.layers = layers | |
| ### Subtract the mean divide by SD | |
| self.inp_norm = lambda inp: normInput(inp, 1/inp_norm_sd, -inp_norm_mean) | |
| def __call__(self, inp): | |
| return self.forward(inp) | |
| def __getitem__(self, i): | |
| return self.layers[i] | |
| def forward(self, inp): | |
| out = self.inp_norm(inp) | |
| for l in self.layers: | |
| out = l(out) | |
| return out |
And that's it - we have a working model. We'll need to figure out how to load weights into this next.
Load weights
PyTorch stores the model weights as a dictionary. Here's an example:
model_filename = "model_conv_medium_2_300_epochs_bs_5.pt"
state_dict = torch.load(model_filename)
state_dict.keys()
---
output:
---
odict_keys(['0.weight', '0.bias', '2.weight', '2.bias', '4.weight', '4.bias', '7.weight', '7.bias', '9.weight', '9.bias', '11.weight', '11.bias', '13.weight', '13.bias'])
---The weights for the model are stored in a dictionary where the keys are {layer_number}.weight or {layer_number}.bias. We save the weights as a JSON file and then write a simple weights loader.
Here's how we can extend the Model class to load the weights from a JSON file:
Load the JSON file into a dictionary and go through the layers one at a time. If weights for that layer exist, pass the weights to that layer's load_state_dict function.
| class Model: | |
| *** skip earlier code *** | |
| def load_weights(self, filename): | |
| with open(filename) as f: | |
| parameters = json.load(f) | |
| keys = [entry for entry in sorted(parameters.keys())] | |
| for i, layer in enumerate(self.layers): | |
| weights = None | |
| bias = None | |
| if f"{i}.weight" in keys: | |
| weights = parameters[f"{i}.weight"] | |
| if f"{i}.bias" in keys: | |
| bias = parameters[f"{i}.bias"] | |
| self.layers[i].load_state_dict(weights, bias) | |
| return self |
Then we need to implement load_state_dict for each layer where we take in the weights and biases and store them for use during inference.
This is how it works for a linear layer:
| class linearLayer(Module): | |
| *** skip earlier code *** | |
| def load_state_dict(self, weights, bias): | |
| self.weights = weights | |
| self.bias = bias |
And this is how it works for a convolution layer:
| class convLayer(Module): | |
| *** skip earlier code *** | |
| def load_state_dict(self, weights, bias): | |
| for i, in_channels in enumerate(weights): | |
| for j, kernels in enumerate(in_channels): | |
| kernel_data = [] | |
| for row in kernels: | |
| kernel_data.append(row) | |
| self.kernels[i][j] = kernel_data | |
| for i, b in enumerate(bias): | |
| self.bias[i] = b |
Validate ESP32 output against output from PyTorch model
It's pretty important to make sure that the model has the same outputs on an ESP32 and on PyTorch.
I wrote test scripts with dummy data for each type of layer. Example test for linear layer:
| from model import linearLayer | |
| def test_linear_layer(weights, biases, input_data): | |
| ### Create an instance of the custom LinearLayer class | |
| custom_layer = linearLayer(weights, biases) | |
| custom_output = custom_layer.forward(input_data) | |
| print("Custom Layer Output: ", custom_output) | |
| ### Create an instance of PyTorch's nn.Linear class | |
| torch_layer = nn.Linear(len(input_data), len(biases)) | |
| with torch.no_grad(): | |
| torch_layer.weight = nn.Parameter(torch.tensor(weights).double()) | |
| torch_layer.bias = nn.Parameter(torch.tensor(biases).double()) | |
| torch_output = torch_layer(torch.tensor(input_data).double()) | |
| print("PyTorch Layer Output: ", torch_output.tolist()) | |
| ### Test case 5 | |
| weights = [[0.5, 0.6, 0.7], [0.8, 0.9, 1.0], [1.1, 1.2, 1.3]] | |
| biases = [0.9, 1.0, 1.1] | |
| input_data = [1.5, 1.6, 1.7] | |
| test_linear_layer(weights, biases, input_data) |
The really nice thing about using MicroPython is that I can import code that is supposed to run on my ESP32 into regular old desktop Python and execute it to validate it. That's what the from model import linearLayer line is doing - importing MicroPython code into desktop Python since it's also perfectly valid desktop Python code. The ability to develop like this makes MicroPython extremely powerful.
Similarly, I picked one datapoint from my dataset and ran it through each model I was testing to make sure the output of the model on MicroPython matches the output of the original PyTorch model for that datapoint.
| from model import * | |
| import random | |
| from single_datapoint_for_testing import * | |
| import time | |
| import sys | |
| if sys.implementation.name != "micropython": | |
| ### I run this on both desktop python and micropython | |
| ### but desktop python doesn't have ticks_ms so I use time.time instead | |
| time.ticks_ms = time.time | |
| def testModel(model): | |
| start_time = time.ticks_ms() | |
| out = model(single_data_point) | |
| end_time = time.ticks_ms() | |
| print(end_time-start_time) | |
| print(out) | |
| testModel(pred_model) |
These tests were especially helpful later when I was writing C optimisations - I just had to rerun the tests to make sure I didn't break anything.
Output from benchmark_model (not the same as our final inference model) used during development that was run on PyTorch:
tensor([-309.3656, 66.2828, -38.7419, -72.0498], grad_fn=<AddBackward0>)
Output from benchmark_model used during development that was run on ESP32:
[-309.3655, 66.28277, -38.74186, -72.04981]
For anyone wondering why the outputs don't match exactly eg. -309.3656 vs -309.3655 - this is because of small errors that are inherent to floating point calculations on GPUs partially due to limitations of floating point numbers and partially due GPUs preferring speed over absolute accuracy so they make optimisations that get "close enough" return values but get them faster.
Here's an example where desktop Python gets a floating point calculation slightly wrong:
>>> 0.7+0.6
1.2999999999999998
Correct answer is:
1.3
So the answer is off by 0.0000000000000002.
But interestingly enough if you do the same calculation but break it up, you get the "correct" answer:
>>> 0.2+0.2+0.2+0.7
1.3
# Even this isn't entirely correct - the actual value in floating point notation is 5854679515581645 / 4503599627370496 which translates to around 1.300000000000000044408920985 - but Python truncates it to 1.3 for display purposes
This is because in floating point operations, the order of operations matters because each number is just an approximate representation of the true value. As you stack more calculations up the errors get larger.
Does it matter? These types of errors can cause issues during training but not so much during inference so for now we can safely ignore it.
Performance optimization
Inference on the benchmark model I was initially using for testing took 194ms. Is that good enough? Maybe. Depends on the use case. But being faster would make this a lot more useful across other embedded use cases and honestly optimization is just fun. Perks of this being on my blog rather than in a product is I can do it just because I want to.
For performance optimization you definitely want to start with a target. Mine was to have inference complete in less than 50ms because that means that you could run inference realtime if you wanted since you collect data every 50ms.
There are three approaches to speeding up inference:
Rewrite specific function calls in C.
eg. the Convolution2D function that convolves a single 2D matrix is a great candidate to be rewritten in C because that has a bunch of loops and rewriting it in C still allows us to retain all of the flexibility of python since we can continue to restructure our model while just calling the function as if it were any other Python function.
There are a few other functions like this that I'll talk about later in the article as well as the exact impact they had on execution time.
The rewritten C functions were compiled as a native module called "mlops" which is a collection of operations useful for machine learning applications. A native module is essentially just a library but compiled into a binary rather than Python code. I upload the "mlops.mpy" library onto the ESP32 like any other MicroPython file and can use it as a module by importing it like any other module with
import mlops
The second approach is to do inference entirely in C and just call the model from MicroPython eg.
c_model.predict(data). This would definitely be the fastest but least flexible - I'd have to rewrite and recompile the C module each time I change model architecture.The third approach is to apply application specific optimizations'. eg. if I was to predict gestures each time a datapoint is collected (every 50ms) then only the last entry of the 1.5s long data recording would change so I just need to update calculations that depend on the last row.
For my use case (a proof of concept on a for-fun blog) I felt the first type of optimization was good enough. I can just predict once every 1.5s and it still works fine.
So now we need to rewrite specific Python functions in C. Let's look at rewriting the the ReLU function in C. As a quick refresher ReLU is a function that replaces all negative elements of a matrix with 0 and leaves the positive elements untouched.
The original Python version is:
| class reluConv(Module): | |
| ### In case anyone is wondering why I've wrapped | |
| ### forward in __call__ - it's because when I am | |
| ### profiling I only want to know the execution time | |
| ### for the full relu call, not for each layer of the recursive call | |
| def __call__(self, output): | |
| return self.forward(output) | |
| def forward(self, output): | |
| if isinstance(output, list): | |
| return [self.forward(i) for i in output] | |
| else: | |
| return max(0, output) |
The C optimized forward function is below.
| STATIC mp_obj_t relu(mp_obj_t input) { | |
| // Check if input is a list | |
| if (mp_obj_is_type(input, &mp_type_list)) { | |
| // Create a new list to hold relu-ed values | |
| mp_obj_t outputs = mp_obj_new_list(0, NULL); | |
| // Convert input list to pointer and get length | |
| mp_obj_list_t *input_c = MP_OBJ_TO_PTR(input); | |
| unsigned int list_len = input_c->len; | |
| for (int i = 0; i < list_len; i++) { | |
| // relu each item in input list | |
| mp_obj_t output = relu(input_c->items[i]); | |
| // append to new list | |
| mp_obj_list_append(outputs, output); | |
| } | |
| return outputs; | |
| } else { | |
| // relu the scalar input | |
| mp_float_t output = mp_obj_get_float(input); | |
| if (output < 0) { | |
| output = 0; | |
| } | |
| return mp_obj_new_float(output); | |
| } | |
| } |
And after we compile that and load the new mlops.mpy library, we rewrite the python class as shown below. This is why I love this approach - it's a really happy medium between having the development speed of Python and the runtime execution speed of C.
| class reluConv(Module): | |
| def __call__(self, output): | |
| return self.forward(output) | |
| def forward(self, output): | |
| return mlops.relu(output) |
Here is a list of performance optimizations. All measurements below were taken at 160MHz clock speed because that's what I started off with and later figured I'll consider the speed increase from the switch to 240MHz during inference as "margin of error" in case things go wrong. I'm glad I did that since I later had to increase model size and therefore it took longer to compute but I was still under my 50ms criteria.
Here are the optimizations and then a chart showing how much time each optimization saved.
194ms was the original inference time
138ms after using the native code emitter in micropython (out of which 56ms/40% of the time is spent in the Convolution2D function)
129ms after specific small performance optimizations (eg. cache values in loops, flatten in C)
74ms after Convolution2D implemented in C
66ms after ReLU forward call implemented in C
49ms after linearLayer matrix multiplication implemented in C
32ms after implementing a matrix summation function in C
21ms after changing the input normalization function to C
These optimizations brought inference time down from 194ms to 23ms @ 160MHz. Inference time was 15ms @ 240MHz.
After I switched to a bigger model inference took 31ms @ 240MHz. If you call this once every 1.5s that's just using 2% of one of the two cores on the ESP32. Plenty of cycles left to do lots of other stuff.
Side Notes
Here's a few small asides.
How many samples are needed?
I looked into whether I really needed 120 samples for each gesture and it does seem like it's roughly correct. Here is model accuracy vs number of samples. These were trained for 2 minutes (50 epochs with one cycle LR scheduler) rather than 13 minutes.
It looks like 75 samples may be enough too but went with higher just to be safe - 96 training samples and 24 validation samples (120 total).
Note: I don't think the slight drop from 75 to 100 is significant, with more runs to sample I feel like it would likely cancel out. From a decision making perspective - 75 or 100 both are probably fine and 100 is safer.
Using coins while I was waiting for the PCB to arrive
After I had the idea to try this and before I received the touch pad PCBs I sent to get made I wanted to carry on with the firmware and software development so I soldered a bunch of coins and put them in a 3D printed casing to start working on the software while I wait for the hardware. It was too big and didn't feel good to "write" on but definitely fun to use coins as touch sensors!
Confidence Threshold
On the ESP32 after the model makes a prediction I only allow it to update the display if it is at least 75% confident that the answer is correct. This does two things:
Prevents it from interpreting ambiguous signals like my palm resting on the touchpad as an alphabet
If it sees an alphabet drawn in a way that confuses the model, it prevents the model from forcing a guess - I found it was often wrong with these types of guesses. It must be quite confident about the prediction to display it on the screen.
Libraries
I tried to look at other libraries like ndarray or upytorch but none of them could do what I needed yet:
uPyTorch doesn't have inference yet
ndarray actually made things slower - code that was pure MicroPython with native emitter that took 138ms to run without ndarray took 179ms after using ndarray.
Appendix - All the other layers
Earlier in the article I mentioned I would show the code for all the other layers - so I've put the code down here along with their C equivalents.
Linear Layer
| class linearLayer(Module): | |
| def __init__(self, weights, bias): | |
| # Initialize weights and bias with given values | |
| self.weights = weights | |
| self.bias = bias | |
| def __call__(self, inp): | |
| return self.forward(inp) | |
| def forward(self, input_data): | |
| intermediate_output = [0 for _ in range(len(self.weights))] | |
| for i in range(len(self.weights)): | |
| for j in range(len(self.weights[0])): | |
| intermediate_output[i] += self.weights[i][j] * input_data[j] | |
| # Add bias | |
| output_data = [intermediate_output[i] + self.bias[i] for i in range(len(self.bias))] | |
| return output_data |
Linear Layer with C Optimization
| class linearLayer(Module): | |
| def __init__(self, weights, bias): | |
| # Initialize weights and bias with given values | |
| self.weights = weights | |
| self.bias = bias | |
| def __call__(self, inp): | |
| return self.forward(inp) | |
| @micropython.native | |
| def forward(self, input_data): | |
| return mlops.linear(input_data, self.weights, self.bias) | |
| def load_state_dict(self, weights, bias): | |
| self.weights = weights | |
| self.bias = bias |
| STATIC mp_obj_t linear(mp_obj_t input_data, mp_obj_t weights, mp_obj_t bias) { | |
| // Convert from mpy lists to pointers | |
| mp_obj_list_t *input_data_c = MP_OBJ_TO_PTR(input_data); | |
| mp_obj_list_t *weights_c = MP_OBJ_TO_PTR(weights); | |
| mp_obj_list_t *weights_row = MP_OBJ_TO_PTR(weights_c->items[0]); | |
| mp_obj_list_t *bias_c = MP_OBJ_TO_PTR(bias); | |
| const int weights_height = weights_c->len;//24, running through 48 inp features 24 outp features in comments as an example | |
| const int weights_width = weights_row->len;//48 | |
| // Convert data to float | |
| float input_data_f[weights_width];//48 | |
| for(int i = 0; i < weights_width; i++){ | |
| input_data_f[i] = mp_obj_get_float(input_data_c->items[i]); | |
| } | |
| float weights_f[weights_height][weights_width];//weights_f[24][48] | |
| mp_obj_list_t *weights_vector; | |
| for(int i = 0; i < weights_height; i++){//24 | |
| weights_vector = MP_OBJ_TO_PTR(weights_c->items[i]); | |
| for(int j = 0; j < weights_width; j++){//48 | |
| weights_f[i][j] = mp_obj_get_float(weights_vector->items[j]); | |
| } | |
| } | |
| // Create array of zeroes | |
| float output[weights_height]; | |
| for(int i = 0; i < weights_height; i++) {//24 | |
| output[i] = mp_obj_get_float(mp_obj_new_float(0.0));//weird workaround for linker bug | |
| } | |
| // Calculate result | |
| for(int i = 0; i < weights_height; i++) {//0 to 24 | |
| for(int j = 0; j < weights_width; j++) {//0 to 48 | |
| output[i] += weights_f[i][j] * input_data_f[j]; | |
| } | |
| } | |
| // Add bias | |
| for(int i = 0; i < weights_height; i++) { | |
| output[i] += mp_obj_get_float(bias_c->items[i]); | |
| } | |
| // Convert to mpy | |
| mp_obj_t output_converted_to_mp[weights_height];//store lists of rows in this | |
| for(int i=0; i < weights_height; i++){ | |
| output_converted_to_mp[i] = mp_obj_new_float(output[i]);//convert row to mp list and store into array | |
| } | |
| return mp_obj_new_list(weights_height, output_converted_to_mp); | |
| } |
ReLU Layer
| class reluConv(Module): | |
| ### In case anyone is wondering why I've wrapped | |
| ### forward in __call__ - it's because when I am | |
| ### profiling I only want to know the execution time | |
| ### for the full relu call, not for each layer of the recursive call | |
| def __call__(self, output): | |
| return self.forward(output) | |
| def forward(self, output): | |
| if isinstance(output, list): | |
| return [self.forward(i) for i in output] | |
| else: | |
| return max(0, output) |
ReLU Layer with C Optimization
| class reluConv(Module): | |
| def __call__(self, output): | |
| return self.forward(output) | |
| def forward(self, output): | |
| return mlops.relu(output) |
| STATIC mp_obj_t relu(mp_obj_t input) { | |
| // Check if input is a list | |
| if (mp_obj_is_type(input, &mp_type_list)) { | |
| // Create a new list to hold relu-ed values | |
| mp_obj_t outputs = mp_obj_new_list(0, NULL); | |
| // Convert input list to pointer and get length | |
| mp_obj_list_t *input_c = MP_OBJ_TO_PTR(input); | |
| unsigned int list_len = input_c->len; | |
| for (int i = 0; i < list_len; i++) { | |
| // relu each item in input list | |
| mp_obj_t output = relu(input_c->items[i]); | |
| // append to new list | |
| mp_obj_list_append(outputs, output); | |
| } | |
| return outputs; | |
| } else { | |
| // relu the scalar input | |
| mp_float_t output = mp_obj_get_float(input); | |
| if (output < 0) { | |
| output = 0; | |
| } | |
| return mp_obj_new_float(output); | |
| } | |
| } |
Dummy Layer
| class Module: | |
| def load_state_dict(self, weights, bias): | |
| pass | |
| class dummyLayer(Module): | |
| def __call__(self, output): | |
| return output |
Flatten Layer
| class Flatten(Module): | |
| def __call__(self, inp): | |
| return self.forward(inp) | |
| @micropython.native | |
| def forward(self, inp): | |
| if not isinstance(inp, (list, tuple)): | |
| return [inp] | |
| result = [] | |
| for i in inp: | |
| if isinstance(i, (list, tuple)): | |
| result.extend(self.forward(i)) | |
| else: | |
| result.append(i) | |
| return result |
Flatten Layer with C Optimization
| class Flatten(Module): | |
| def __call__(self, inp): | |
| return self.forward(inp) | |
| @micropython.native | |
| def forward(self, inp): | |
| return mlops.flatten(inp) |
| STATIC mp_obj_t flatten(mp_obj_t input) { | |
| // Check if input is a list or tuple | |
| if (mp_obj_is_type(input, &mp_type_list) || mp_obj_is_type(input, &mp_type_tuple)) { | |
| // Create a new list to hold flattened values | |
| mp_obj_t outputs = mp_obj_new_list(0, NULL); | |
| // Convert input list to pointer and get length | |
| mp_obj_list_t *input_c = MP_OBJ_TO_PTR(input); | |
| unsigned int list_len = input_c->len; | |
| for (int i = 0; i < list_len; i++) { | |
| // Check if item is a list or tuple | |
| if (mp_obj_is_type(input_c->items[i], &mp_type_list) || mp_obj_is_type(input_c->items[i], &mp_type_tuple)) { | |
| // Flatten the item and extend the output list | |
| mp_obj_t flattened = flatten(input_c->items[i]); | |
| mp_obj_list_t *flattened_c = MP_OBJ_TO_PTR(flattened); | |
| for (int j = 0; j < flattened_c->len; j++) { | |
| mp_obj_list_append(outputs, flattened_c->items[j]); | |
| } | |
| } else { | |
| // Append the item to the output list | |
| mp_obj_list_append(outputs, input_c->items[i]); | |
| } | |
| } | |
| return outputs; | |
| } else { | |
| // Return the scalar input as a list | |
| return mp_obj_new_list(1, &input); | |
| } | |
| } |
Input Normalization
| @micropython.native | |
| def normInput(matrix, multiplier, adder): | |
| for i, element in enumerate(matrix): | |
| if isinstance(element, (list, array.array)): | |
| # If the current element is a list, recursively call the function on this element | |
| normInput(element, multiplier, adder) | |
| else: | |
| # If the current element is not a list, multiply it by the multiplier and add the adder | |
| matrix[i] = (element + adder) * multiplier | |
| return matrix |
Input Normalization with C Optimization
| @micropython.native | |
| def normInput(matrix, multiplier, adder): | |
| return mlops.norm_input(multiplier, adder) |
| STATIC mp_obj_t norm_input(mp_obj_t input, mp_obj_t multiplier, mp_obj_t adder) { | |
| // Check if input is a list | |
| if (mp_obj_is_type(input, &mp_type_list)) { | |
| // Create a new list to hold normalized values | |
| mp_obj_t outputs = mp_obj_new_list(0, NULL); | |
| // Convert input list to pointer and get length | |
| mp_obj_list_t *input_c = MP_OBJ_TO_PTR(input); | |
| unsigned int list_len = input_c->len; | |
| for (int i = 0; i < list_len; i++) { | |
| // relu each item in input list | |
| mp_obj_t normalized = norm_input(input_c->items[i], multiplier, adder); | |
| // append to new list | |
| mp_obj_list_append(outputs, normalized); | |
| } | |
| return outputs; | |
| } else { | |
| // relu the scalar input | |
| float input_f = mp_obj_get_float(input); | |
| float multiplier_f = mp_obj_get_float(multiplier); | |
| float adder_f = mp_obj_get_float(adder); | |
| return mp_obj_new_float((input_f+adder_f)*multiplier_f); | |
| } | |
| } |
Conv2D Layer
| class convLayer(Module): | |
| def __init__(self, in_channels, out_channels, kernel_size: tuple, stride): | |
| self.kernels = [[None for _ in range(in_channels)] for _2 | |
| in range(out_channels)] | |
| self.bias = [None for _ in range(out_channels)] | |
| self.stride = stride | |
| self.kernel_size = kernel_size | |
| def __call__(self, images): | |
| conv_func = convolution2D | |
| output_data = [] | |
| for out_channel in range(len(self.kernels)): | |
| channel_output = [] | |
| for in_channel, image in enumerate(images): | |
| channel_output.append(conv_func(image, self.kernels[out_channel][in_channel],self.stride)) | |
| ### Matrix addition operation | |
| ### sum up all the different outputs for each channel | |
| summed_channel_output = sumMatrices(channel_output) | |
| ### Add the bias for each channel to each element | |
| ### in the output matrix for that channel | |
| biased_channel_output = addBias(summed_channel_output, self.bias[out_channel]) | |
| output_data.append(biased_channel_output) | |
| return output_data | |
| def convolution2D(image, kernel, stride): | |
| ### Convolve single 2D matrix with kernel using stride | |
| ### returns a single 2D matrix | |
| ### Get dimensions of the input matrix (image) and the kernel | |
| image_height, image_width = int(len(image)), int(len(image[0])) | |
| kernel_height, kernel_width = int(len(kernel)), int(len(kernel[0])) | |
| ### Calculate dimensions of the output image | |
| output_height = (image_height - kernel_height) // stride + 1 | |
| output_width = (image_width - kernel_width) // stride + 1 | |
| ### Initialize output image with zeros | |
| output = [[0]*output_width for _ in range(output_height)] | |
| ### Iterate over the image | |
| for i in range(0, image_height, stride): | |
| ### optimization | |
| output_y = i//stride | |
| for j in range(0, image_width, stride): | |
| ### optimization | |
| output_x = j//stride | |
| ### Check if the kernel can fit in the image | |
| if i + kernel_height <= image_height and j + kernel_width <= image_width: | |
| ### Apply the kernel to the image | |
| for m in range(kernel_height): | |
| i_plus_m = i+m | |
| for n in range(kernel_width): | |
| output[output_y][output_x] += image[i_plus_m][j+n] * kernel[m][n] | |
| return output |
Conv2D Layer with C optimization
| class convLayer(Module): | |
| def __init__(self, in_channels, out_channels, kernel_size: tuple, stride): | |
| self.kernels = [[None for _ in range(in_channels)] for _2 | |
| in range(out_channels)]# Start with no kernels | |
| self.bias = [None for _ in range(out_channels)]# Start with no kernels | |
| self.stride = stride | |
| self.kernel_size = kernel_size | |
| @micropython.native | |
| def __call__(self, images): | |
| conv_func = mlops.conv2d | |
| output_data = [] | |
| for out_channel in range(len(self.kernels)): | |
| channel_output = [] | |
| for in_channel, image in enumerate(images): | |
| channel_output.append(conv_func(image, self.kernels[out_channel][in_channel],self.stride)) | |
| biased_channel_output = sumMatricesAddBias(channel_output, self.bias[out_channel]) | |
| output_data.append(biased_channel_output) | |
| return output_data | |
| def load_state_dict(self, weights, bias): | |
| for i, in_channels in enumerate(weights): | |
| for j, kernels in enumerate(in_channels): | |
| kernel_data = [] | |
| for row in kernels: | |
| kernel_data.append(row) | |
| self.kernels[i][j] = kernel_data | |
| for i, b in enumerate(bias): | |
| self.bias[i] = b | |
| @micropython.native | |
| def sumMatricesAddBias(matrices, bias=0.0): | |
| return mlops.sum_matrices(matrices, bias) |
| STATIC mp_obj_t conv2d(mp_obj_t image, mp_obj_t kernel, mp_obj_t stride) { | |
| // Convert from mpy lists to pointers | |
| mp_obj_list_t *image_c = MP_OBJ_TO_PTR(image); | |
| mp_obj_list_t *image_row = MP_OBJ_TO_PTR(image_c->items[0]); | |
| mp_obj_list_t *kernel_c = MP_OBJ_TO_PTR(kernel); | |
| mp_obj_list_t *kernel_row = MP_OBJ_TO_PTR(kernel_c->items[0]); | |
| // Convert from mpy object to c | |
| const int stride_c = mp_obj_get_int(stride); | |
| const size_t image_height = image_c->len; | |
| const size_t image_width = image_row->len; | |
| const size_t kernel_height = kernel_c->len; | |
| const size_t kernel_width = kernel_row->len; | |
| const int output_height = (image_height - kernel_height) / stride_c + 1; | |
| const int output_width = (image_width - kernel_width) / stride_c + 1; | |
| // Create output array of zeroes | |
| float output[output_height][output_width]; | |
| for(int i = 0; i < output_width; i++) { | |
| for(int j = 0; j < output_height; j++){ | |
| output[j][i] = 0.0; | |
| } | |
| } | |
| // Convolve | |
| int output_y; | |
| int output_x; | |
| int i_plus_m; | |
| for(int i = 0; i < image_height; i+= stride_c){ | |
| output_y = i/stride_c; | |
| for(int j=0; j < image_width; j+= stride_c){ | |
| output_x = j/stride_c; | |
| if (i + kernel_height <= image_height && j + kernel_width <= image_width) { | |
| // code to execute if condition is true | |
| for(int m=0; m < kernel_height; m++){ | |
| i_plus_m = i+m; | |
| image_row = MP_OBJ_TO_PTR(image_c->items[i_plus_m]); | |
| kernel_row = MP_OBJ_TO_PTR(kernel_c->items[m]); | |
| for(int n=0; n < kernel_width; n++){ | |
| output[output_y][output_x] += mp_obj_get_float(image_row->items[j+n]) * mp_obj_get_float(kernel_row->items[n]); | |
| } | |
| } | |
| } | |
| } | |
| } | |
| // Convert result calculated above to mpy compatible object | |
| mp_obj_t temp_row[output_width];//store rows in this | |
| mp_obj_t output_converted_to_mp[output_height];//store lists of rows in this | |
| for(int i=0; i < output_height; i++){ | |
| for(int j = 0; j < output_width; j++){ | |
| // first convert all values to float | |
| temp_row[j] = mp_obj_new_float(output[i][j]); | |
| } | |
| output_converted_to_mp[i] = mp_obj_new_list(output_width, temp_row);//convert row to mp list and store into array | |
| } | |
| return mp_obj_new_list(output_height, output_converted_to_mp); | |
| } | |
| STATIC mp_obj_t sum_matrices(mp_obj_t matrices, mp_obj_t bias) { | |
| // Convert from mpy lists to pointers and mpy float to c float | |
| float bias_f = mp_obj_get_float(bias); | |
| mp_obj_list_t *matrices_c = MP_OBJ_TO_PTR(matrices); | |
| mp_obj_list_t *first_matrix = MP_OBJ_TO_PTR(matrices_c->items[0]); | |
| mp_obj_list_t *first_matrix_first_row = MP_OBJ_TO_PTR(first_matrix->items[0]); | |
| int num_matrices = matrices_c->len; | |
| int matrix_height = first_matrix->len; | |
| int matrix_width = first_matrix_first_row->len; | |
| // Initialize result matrix with zeroes | |
| float result[matrix_height][matrix_width]; | |
| for(int i = 0; i < matrix_height; i++){ | |
| for (int j=0; j < matrix_width; j++){ | |
| result[i][j] = bias_f; | |
| } | |
| } | |
| mp_obj_list_t *matrix; | |
| mp_obj_list_t *matrix_row; | |
| for(int n = 0; n < num_matrices; n++){ | |
| matrix = MP_OBJ_TO_PTR(matrices_c->items[n]); | |
| for(int i=0; i < matrix_height; i++){ | |
| matrix_row = MP_OBJ_TO_PTR(matrix->items[i]); | |
| for(int j = 0; j < matrix_width; j++){ | |
| result[i][j] += mp_obj_get_float(matrix_row->items[j]); | |
| } | |
| } | |
| } | |
| // Convert result calculated above to mpy compatible object | |
| mp_obj_t temp_row[matrix_width];//store rows in this | |
| mp_obj_t output_converted_to_mp[matrix_height];//store lists of rows in this | |
| for(int i=0; i < matrix_height; i++){ | |
| for(int j = 0; j < matrix_width; j++){ | |
| // first convert all values to float | |
| temp_row[j] = mp_obj_new_float(result[i][j]); | |
| } | |
| output_converted_to_mp[i] = mp_obj_new_list(matrix_width, temp_row);//convert row to mp list and store into array | |
| } | |
| return mp_obj_new_list(matrix_height, output_converted_to_mp); | |
| } |